About PastML
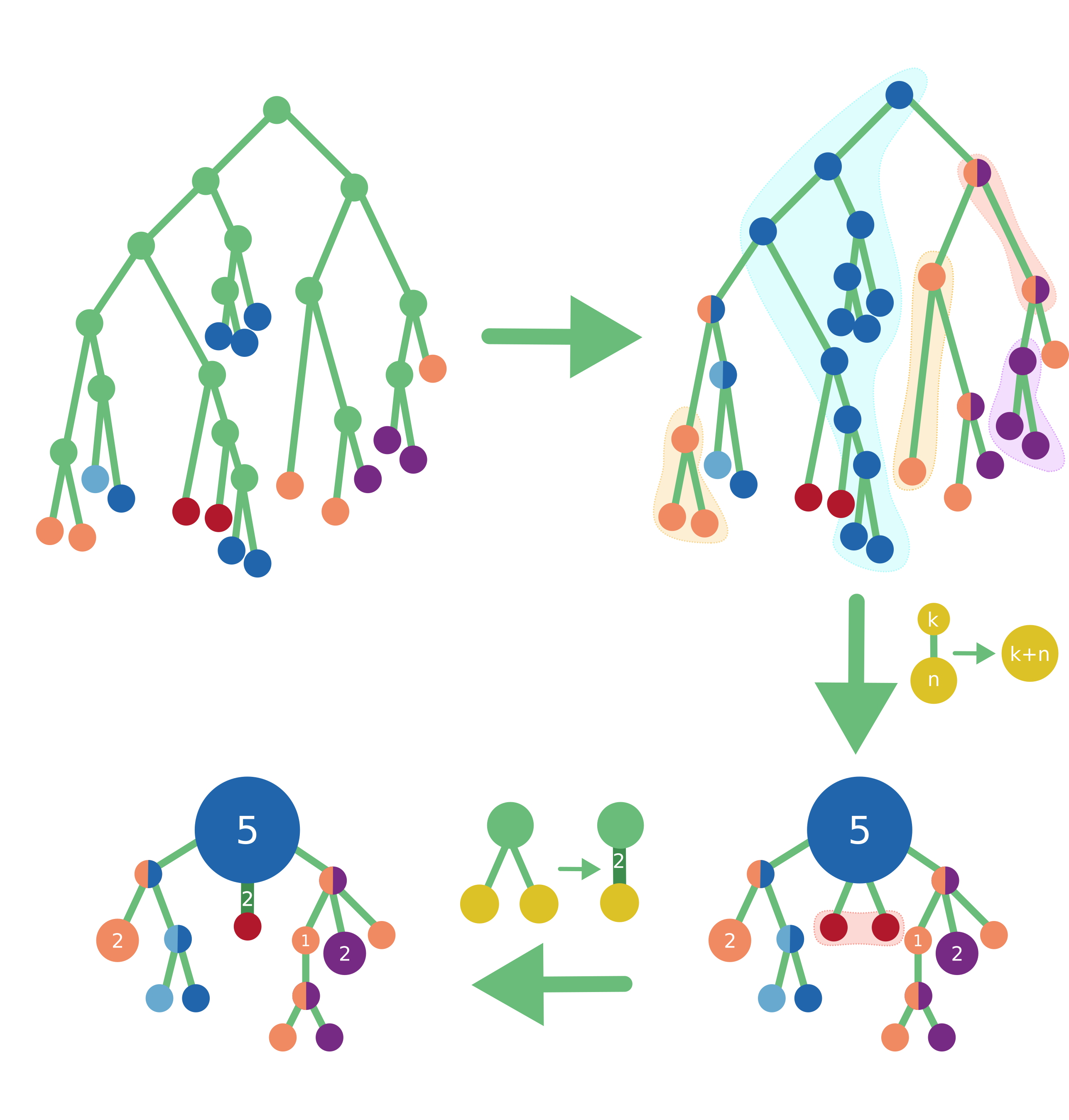
PastML infers ancestral characters on a rooted phylogenetic tree with annotated tips, using maximum likelihood or parsimony. The result is then visualised as a zoomable html map.
To learn more about how PastML works, please visit our Help page.
To learn even more, to cite PastML, or to see the data and simulations described in the article, see here.
To install PastML locally or use it via docker, follow the installation instructions.
PastML source code is available on github.
Cite PastML
Ishikawa SA, Zhukova A, Iwasaki W, Gascuel O (2019) A Fast Likelihood Method to Reconstruct and Visualize Ancestral Scenarios. Molecular Biology and Evolution, msz131.
Try it now
You need two files to run PastML:
Run PastMLExample data
If you prefer, try PastML on our example.
HIV-1A in Albania
This data set comes from the study by Chevenet et al., 2013, which adopted the data from Salemi et al., 2008, and includes:- a rooted HIV-1A tree with 152 tips,
- a comma-separated annotation file describing the country of sampling for each tree tip.
Check out the visualisation of the Albanian data phylogeography: compressed visualisation and full tree visualisation.
Download data


